Input data and parameters
Input
| Analysis date: | Wed Sep 02 10:21:43 CEST 2015 |
| BAM file: | /data/qualimap_release_data/counts/kidney.bam |
| Counting algorithm: | uniquely-mapped-reads |
| GTF file: | /data/qualimap_release_data/annotations/human.64.gtf |
| Paired-end sequencing: | no |
| Protocol: | non-strand-specific |
| Sorting performed: | no |
Summary
Reads alignment
| Number of mapped reads: | 10,135,485 |
| Total number of alignments: | 10,135,485 |
| Number of secondary alignments: | 0 |
| Number of non-unique alignments: | 4,464,055 |
| Aligned to genes: | 4,255,142 |
| Ambiguous alignments: | 85,895 |
| No feature assigned: | 1,330,393 |
| Not aligned: | 0 |
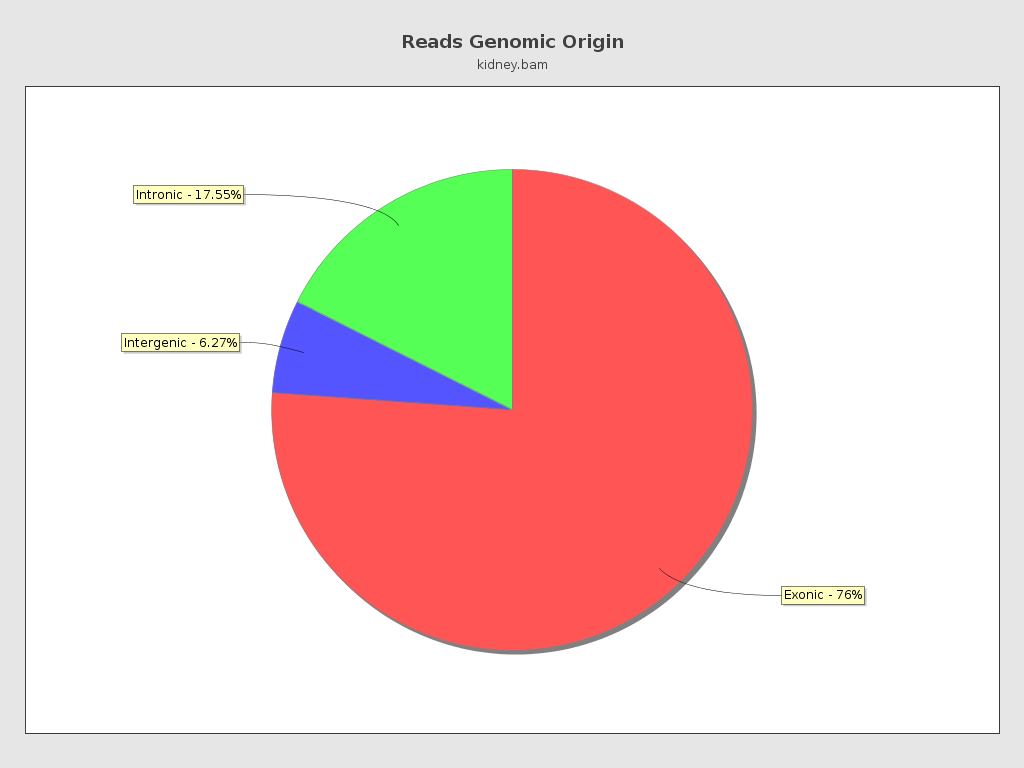
Reads genomic origin
| Exonic: | 4,255,142 / 76.18% |
| Intronic: | 980,432 / 17.55% |
| Intergenic: | 349,961 / 6.27% |
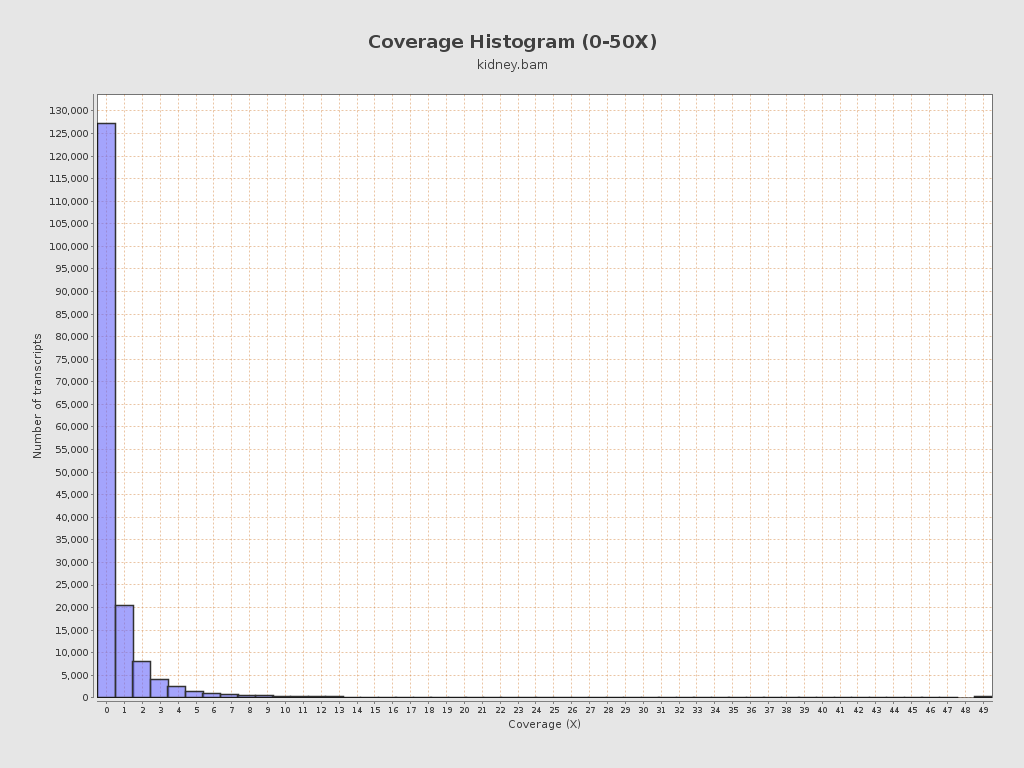
Transcript coverage profile
| 5' bias: | 0.71 |
| 3' bias: | 0.69 |
| 5'-3' bias: | 1.1 |
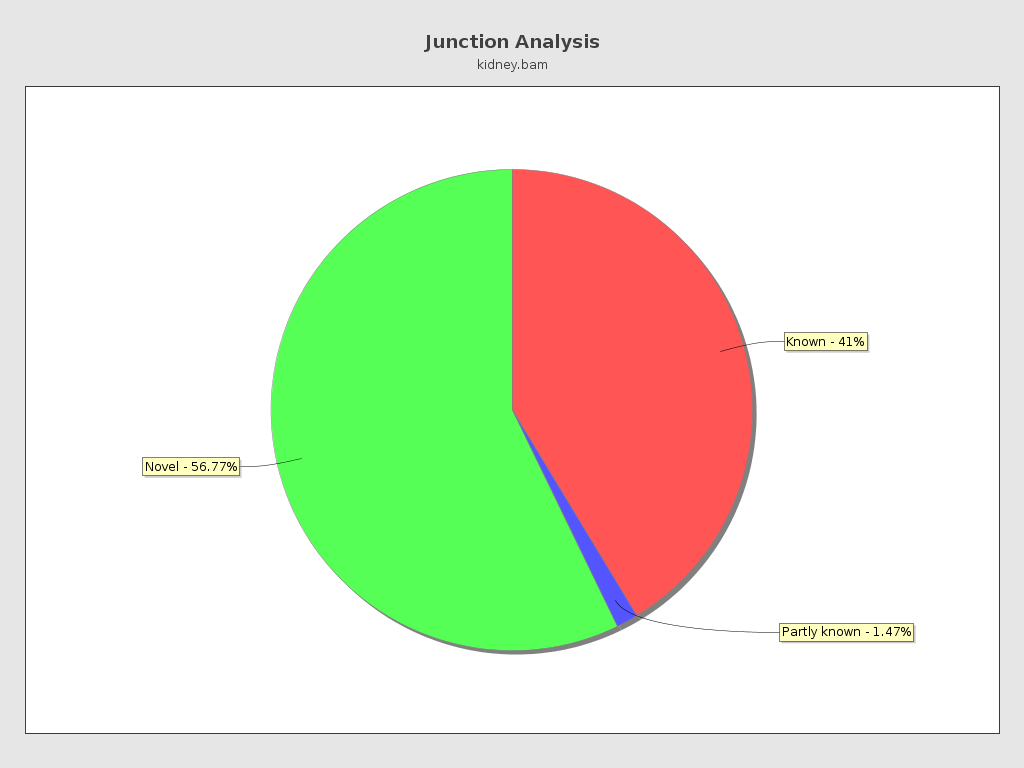
Junction analysis
| Reads at junctions: | 86,449 |
| GCCT | 3.99% |
| ACCT | 3.93% |
| AGGT | 3.54% |
| AGGC | 3.54% |
| AGCT | 2.44% |
| TCCT | 2.24% |
| AGGA | 2.15% |
| ATCT | 2.13% |
| AGAT | 1.76% |
| TTCT | 1.52% |
| CCCT | 1.43% |

.png)
.png)
.png)